
Coenzyme F430 from Methanogenic Archaea
ETH-Zurich | Department of Chemistry | Laboratory of Organic Chemistry | Jaun Group Homepage
Research
Methanogenic archaea are strictly anaerobic microorganisms living in a variety of oxygen-free environments such as sediments of lakes, swamps, rice fields, the rumen of cows, the intestine of termites or the anaerobic stage of sewage plants. They gain their energy for ATP synthesis by producing methane from either CO2/H2 or from acetate.
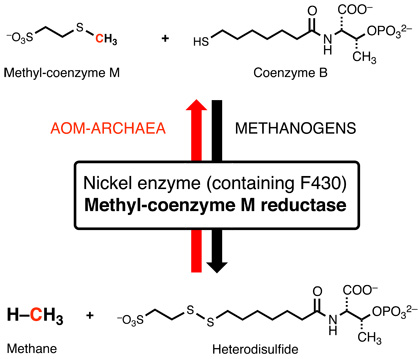
Recently discovered archaea (ANME-I to -III) live in communities with sulfate reducing bacteria near methane seeps and on methane-hydrate deposits in the oceans. They oxidize methane anaerobically (AOM) using sulfate as the final electron acceptor and contain enzymes that are highly homologous to methyl coenzyme M reductase. According to the hypothesis of "Reverse Methanogenesis" they use essentially the same enzyme and pathways to catalyze methane functionalization by reversing the process of methanogenesis. We have recently been able to prove that MCR from a methanogen is indeed able to functionalize methane at rates comparable to those estimated for the in vivo process in ANME archaea.This means that the mechanisms of MCR in methane formation by methanogens and in C-H activation in AOM are one and the same.
The key enzyme for both processes is methyl coenzyme M reductase (MCR). Its active site contains the hydroporphinoid nickel complex coenzyme F430. The nickel center of coenzyme F430 catalyzes the the unusual reaction shown below:
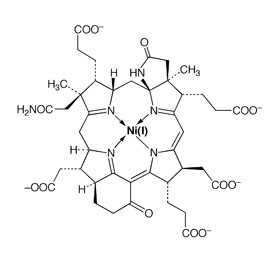
Coenzyme
F430 in the
catalytically active Ni(I) valence form
The elucidation of the still unknown catalytic mechanism of MCR is the focus of our scientific interests.
Using stable isotopes we investigate isotope exchange and isotope effects during catalysis by the active enzyme with labeled substrates and substrate analogs. The coordination environment of the nickel in different enzyme states can be deduced from EPR/ENDOR studies with labeled substrates. We also study the free coenzyme and its derivatives as models for its catalytic function in the enzyme. To this purpose, we use physical-organic tools such as electroanalytical chemistry, spectro-electrochemistry, NMR- and EPR spectroscopy, kinetics, isotope labeling and kinetic isotope effects. An important aspect of our work towards understanding the catalytic mechanism is the investigation of the relationship between the structure and conformation of the macrocyclic ligand and the reactivity of the nickel center in its different oxidation-, spin- and coordination states. Quantum chemical calculations (DFT) help to interpret the spectroscopic data and analyse the electronic structure of F430.
Collaborations: R.K. Thauer (MPI for Terrestrial Microbiology, Marburg, D), F. Widdel (MPI for Marine Microbiology, Bremen, DE), M. Reiher (Physical Chemistry ETH), J. Harmer (Oxford University, GB), M.K. Johnson (University of Georgia-Athens, USA), E.C. Duin (Auburn University, USA).
Jaun
Group Homepage
Editor: B. Jaun |![]() | Last update: 25.2.2013
| Last update: 25.2.2013
!!! This document is stored in the ETH Web archive and is no longer maintained !!!